Brainsight¶
This module provides import and export functions for the Brainsight TMS Navigator software.
SimNIBS requires an individual T1-NIfTI to be used during neuronavigation. This T1 scan should be the preprocessed T1 image that is created during the SimNIBS headmeshing procedure. Although DICOMS can be used for the Brainsight neuronavigation, no import/export to SimNIBS is supported, due to (possible) different coordinate systems.
Brainsight provides two main ways of storing positions/orientations: Targets and Samples. Targets represent planned locations for stimulation, and Samples represent the actual locations that were stimulated. Both are exported in a single .txt file. As several coordinate systems are supported for export and import by Brainsight, please see the following notes regarding which of them are suited for data exchange with SimNIBS.
How to use¶
Exporting from Brainsight¶
With Brainsight 2.5.2 and earlier, choose the NIfTI:Aligned coordinate system when exporting.
With Brainsight 2.5.3 and later, choose the NIfTI:Q:xxx coordinate system when exporting. (The XXX will be a string version of the intent code, and will be one of: Aligned, Scanner, MNI-152, Talairach, or Other-Template.)
Importing to SimNIBS¶
simnibs.brainsight.read(fn) reads Brainsight .txt files and returns two simnibs.TMSLIST() objects: One for Targets and one for Samples. The conversion of the different coil axes definitions is performed automatically.
simnibs.TMSLIST()¶from simnibs import sim_struct, brainsight
s = sim_struct.SESSION()
fn = "exported_data.txt"
tms_list_targets, tms_list_samples = brainsight().read(fn) # read all Targets and Samples from file and return as TMSLIST() each
s.add_tmslist(tms_list_targets)
tms_list.pos[0].name # <- name is filled with data from .txt.
Exporting from SimNIBS¶
simnibs.brainsight.write(obj, fn) writes a .txt file that is compatible for Brainsight import. The conversion between the different coil axes definitions is performed automatically.
import numpy as np
from simnibs import sim_struct, opt_struct, brainsight
fn = "precomuted_coilpos.xml"
### export from TMSLIST
tmslist = sim_struct.TMSLIST()
tmslist.add_position()
# ... define (multiple) positions ...
brainsight().write(tmslist, fn)
### export from POSITION
pos = sim_struct.POSITION()
pos.matsimnibs = ...
brainsight().write(pos, fn)
### export from np.ndarray / matsimnibs
opt = opt_struct.TMSoptimize()
# ... prepare optmization ...
opt_mat = opt.run() # get optimal position
brainsight().write(np.squeeze(opt_mat), fn)
Notes¶
The same anatomical scan has to be used for Brainsight and SimNIBS.
The same coil model has to be used for field simulations and for real stimulation.
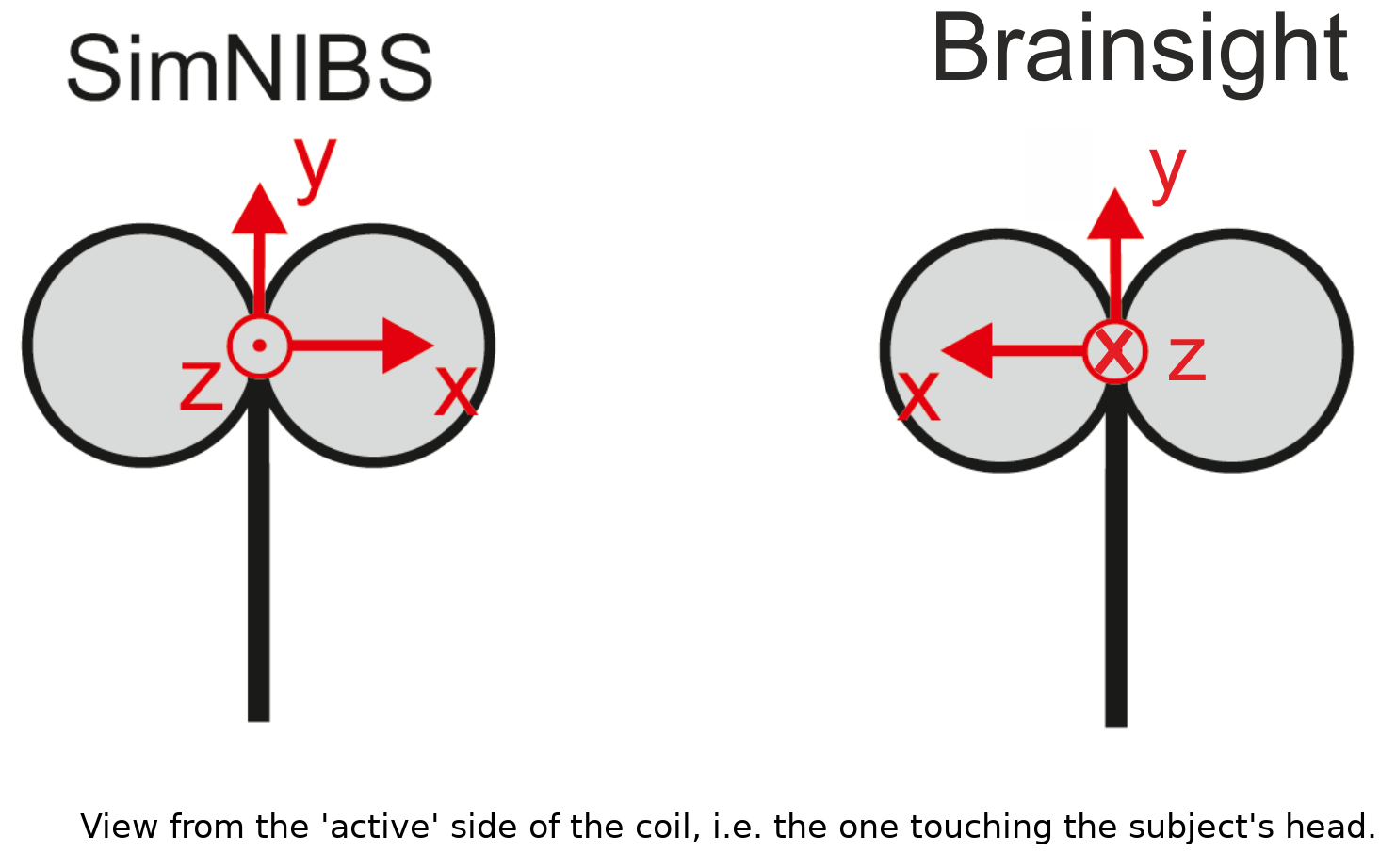
Coordinate systems used to define coil axes for SimNIBS and Brainsight: