Fixing the Affine Registration¶
Note
This tutorial applies only when setting init_type = "atlas" in the charm configuration file (settings.ini).
When encountering spurious segmentation results, we have found that this can often be traced back to a suboptimal affine registration between the structural scan(s) and the atlas. Identifying what exactly is causing the segmentation to fail can be a bit tricky (at least initially) so here we provide a few examples showing the type of errors that one might encounter and how they can be alleviated by modifying the settings of charm. Charm writes a settings.ini inside the m2m-folder of the subject. Make a copy of this and then modify it as described below. This way, different changes can be made for individual subjects. To use the modified settings file, pass it using the –usesettings argument when running charm.
Here we use the visualizations provided by charm in the charm_report.html document, specifically, the Coregistered template and Tissue labels overlays.
1. Bad Rotation¶
In the final segmentation, we see what appears to be a downwards rotation around the x-axis. We can fix this by changing the default value of affine_rotations (which is [-7, -3.5, 0, 3.5, 7]) to for example affine_rotations = [0], thus restricting the rotational initializations around the x-axis.
affine_rotations are specified as a list of initial rotations in degrees.
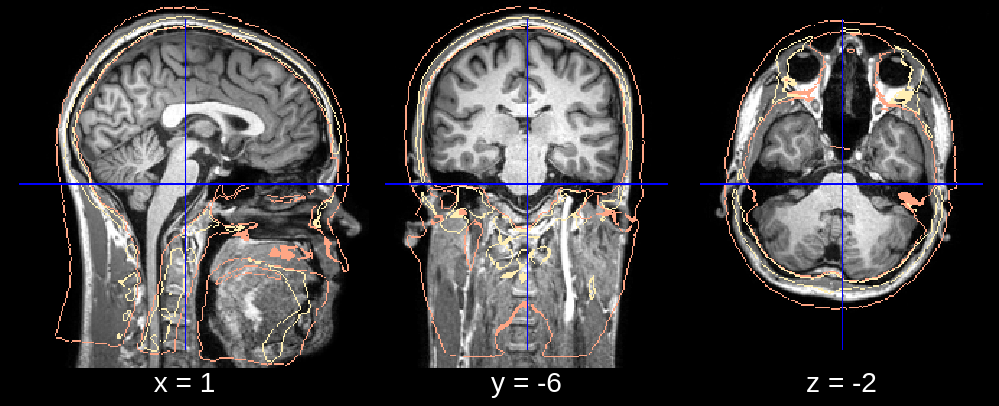
Fig 1.1. Affine (rotation issue).¶
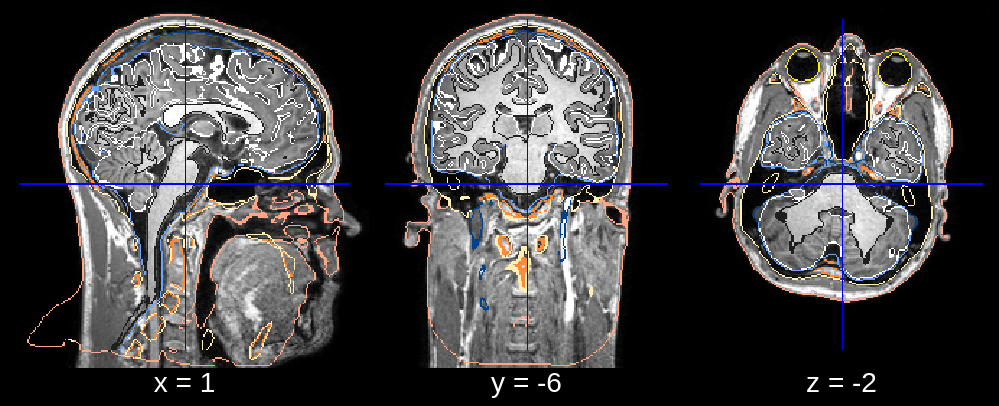
Fig 1.2. Final segmentation (rotation issue).¶
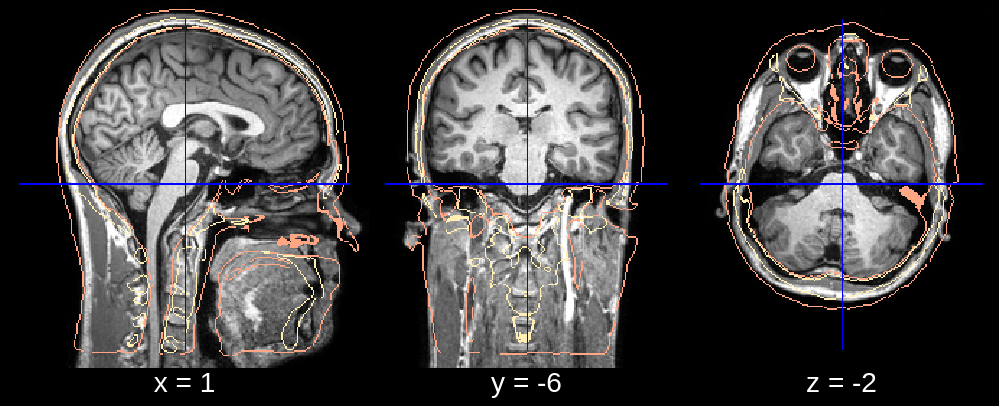
Fig 1.3. Affine (rotation fixed).¶
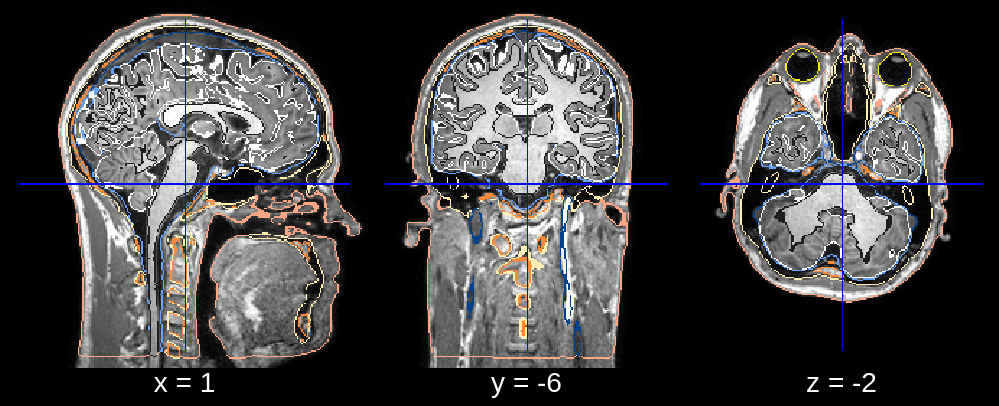
Fig 1.4. Final segmentation (rotation fixed).¶
1. Bad Scaling¶
In the final segmentation, we see a clear dent in the frontal part of the head. Looking at the affine registration it seems that the atlas was scaled down too much. We can fix this by changing the default value of affine_scales (which is [[0.85, 0.85, 0.85], [0.9, 0.9, 0.85], [0.95, 0.95, 0.85]]) to for example affine_scales = [[0.9, 1.0, 0.85], [0.95, 1.0, 0.85]] where we make the initializations of the scaling (along the x- and y-axis) larger.
affine_scales is specified as a list of initial scalings in along each axis, i.e., [x, y, z].
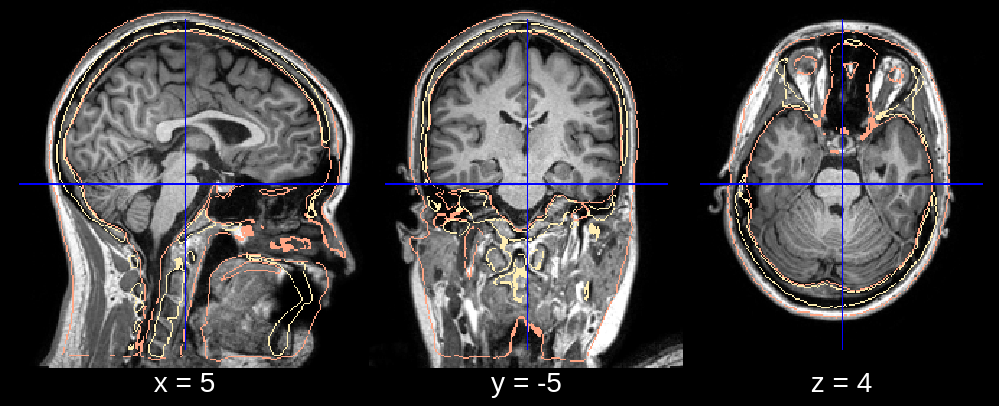
Fig 2.1. Affine (scale issue).¶
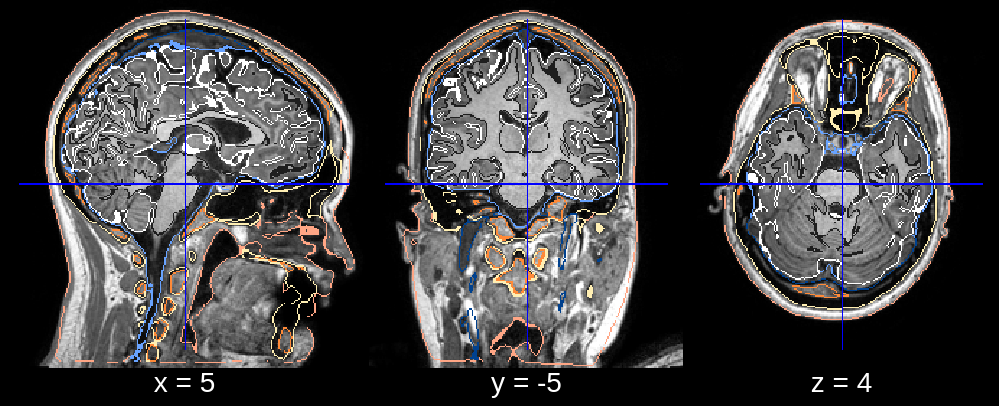
Fig 2.2. Final segmentation (scaling issue).¶
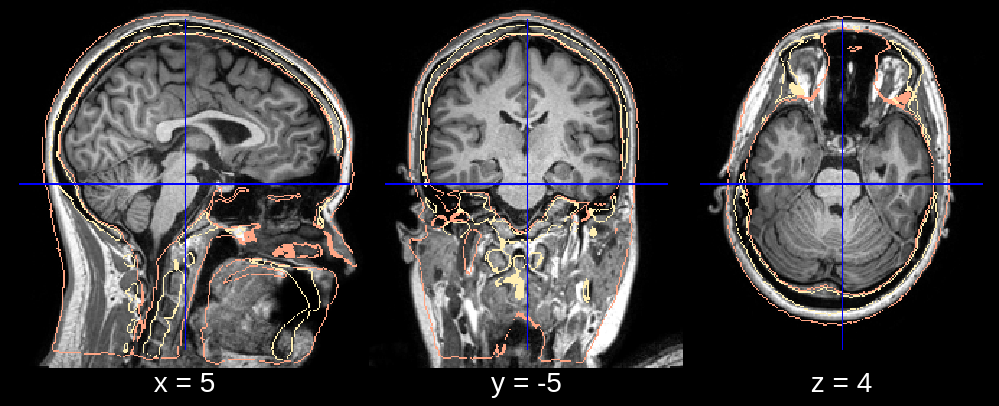
Fig 2.3. Affine (scaling fixed).¶
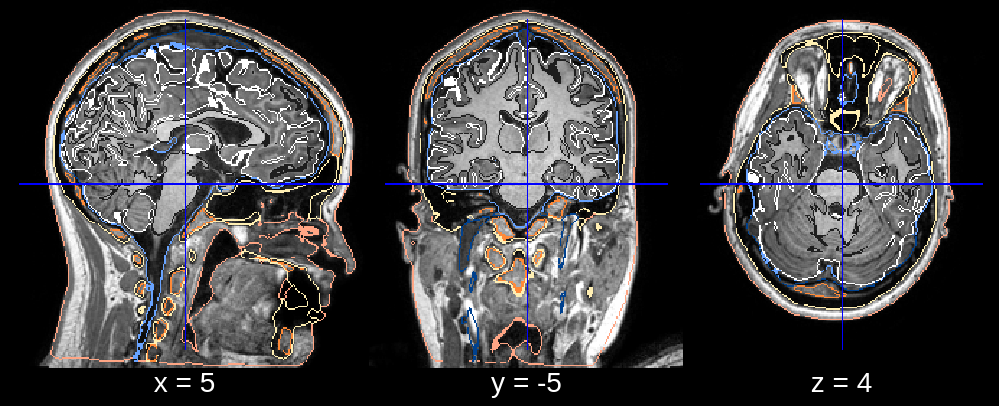
Fig 2.4. Final segmentation (scaling fixed).¶
3. Bad Neck Deformation¶
In the final segmentation, we see that the neck is deformed too far back. This can be fixed by restricting the posterior search bound of the neck deformation. Specifically, we can replace the default value of neck_search_bounds (which is [-0.3, 0.1]) with neck_search_bounds = [0, 0.2] to prevent posterior deformation and allow slightly more deformation in the anterior direction.
neck_search_bounds is specified as a list with a posterior and an anterior search bound.
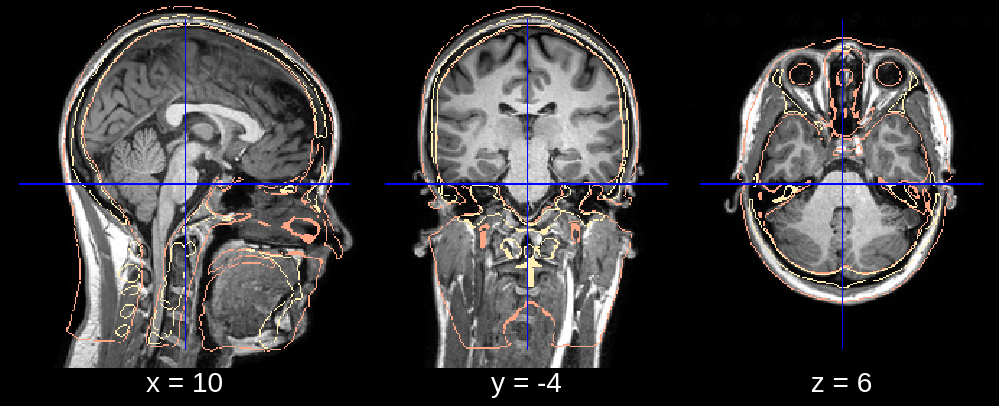
Fig 3.1. Affine (neck deformation issue).¶
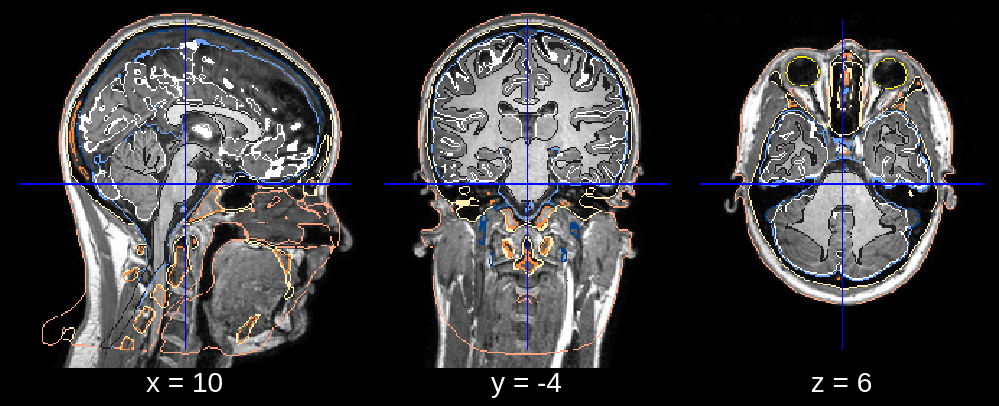
Fig 3.2. Final segmentation (neck deformation issue).¶
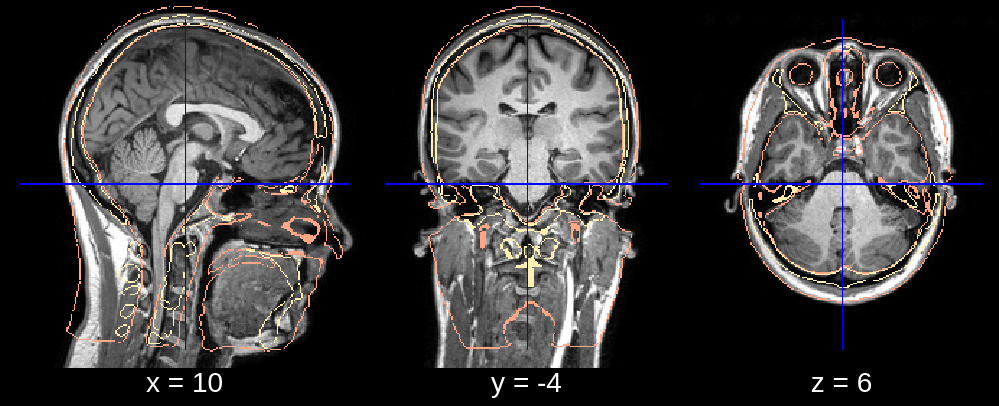
Fig 3.3. Affine (neck deformation fixed). Since the neck deformation is performed after the affine registration, there is no visual effect of this, but the result can be appreciated in the final segmentation (fig. 3.4).¶
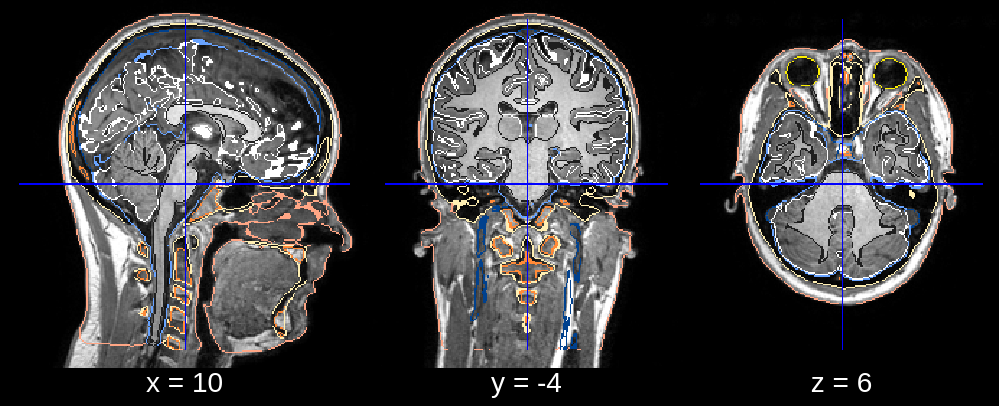
Fig 3.4. Final segmentation (neck deformation fixed)¶
